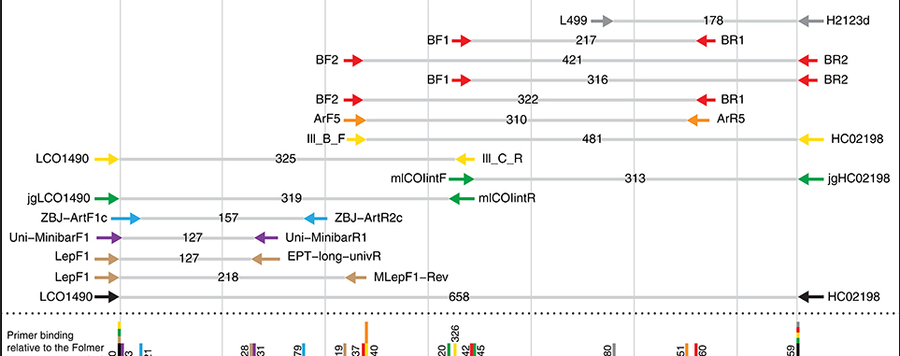
Validation and development of COI metabarcoding primers for freshwater macroinvertebrate bioassessment
A central challenge in the present era of biodiversity loss is to assess and manage human impacts on freshwater ecosystems. Macroinvertebrates are an important group for such bioassessments as many taxa show specific responses to environmental conditions. However, generating accurate macroinvertebrate inventories based on primarily larval morphology is difficult and error-prone. Here, DNA metabarcoding provides new opportunities. Its potential to accurately identify invertebrates in bulk samples to the species level has been demonstrated in several case studies. However, DNA based identification is often limited by primer bias, potentially leading to taxa in the sample remaining undetected. Thus, the success of DNA metabarcoding as an emerging technique for bioassessment critically relies on carefully evaluated primers. We used the R package PrimerMiner to obtain and process cytochrome c oxidase I (COI) sequence data for the 15 globally most relevant freshwater invertebrate groups for stream assessment. Using these sequence alignments, we developed four primer combinations optimized for freshwater macroinvertebrates. All primers were evaluated by sequencing 10 mock community samples, each consisting of 52 freshwater invertebrate taxa. Additionally, popular metabarcoding primers from the literature and the developed primers were tested in silico against these 15 relevant invertebrate groups. The developed primers varied in amplification efficiency and the number of detected taxa, yet, all detected more taxa than standard “Folmer” barcoding primers. Two new primer combinations showed even more consistent amplification than a previously tested ribosomal marker (16S) and detected all 42 insect taxa present in the mock community samples. In silico evaluation revealed critical design flaws in some commonly used primers from the literature. We demonstrate a reliable strategy to develop optimized primers using the tool PrimerMiner. The developed primers detected almost all taxa present in the mock samples, and we argue that high base degeneracy is necessary to decrease primer bias as confirmed by experimental results and in silico primer evaluation. We further demonstrate that some primers currently used in metabarcoding studies may not be suitable for amplification of freshwater macroinvertebrates. Therefore, careful primer evaluation and more region/ecosystem specific primers are needed before DNA metabarcoding can be used for routine bioassessment of freshwater ecosystems.






