Testing the Global Malaise Trap Program – How well does the current barcode reference library identify flying insects in Germany?
Biodiversity patterns are inherently complex and difficult to comprehensively assess. Yet, deciphering shifts in species composition through time and space are crucial for efficient and successful management of ecosystem services, as well as for predicting change. To better understand species diversity patterns, Germany participated in the Global Malaise Trap Program, a world-wide collection program for arthropods using this sampling method followed by their DNA barcode analysis. Traps were deployed at two localities: “Nationalpark Bayerischer Wald” in Bavaria, the largest terrestrial Natura 2000 area in Germany, and the nature conservation area Landskrone, an EU habitats directive site in the Rhine Valley. Arthropods were collected from May to September to track shifts in the taxonomic composition and temporal succession at these locations.
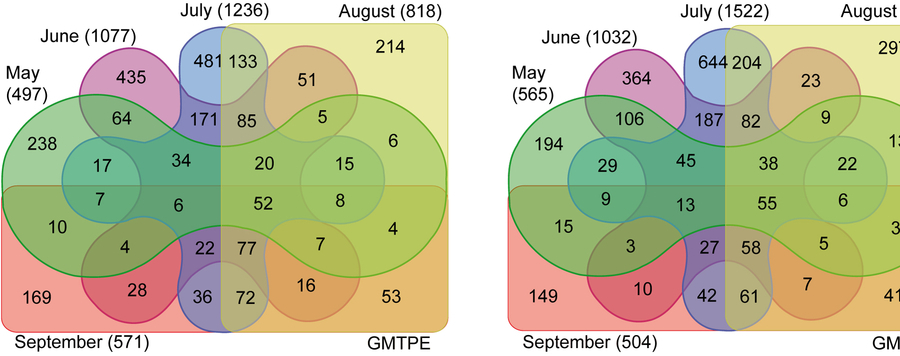
In total, 37,274 specimens were sorted and DNA barcoded, resulting in 5,301 different genetic clusters (BINs, Barcode Index Numbers, proxy for species) with just 7.6% of their BINs shared. Accumulation curves for the BIN count versus the number of specimens analyzed suggest that about 63% of the potential diversity at these sites was recovered with this single season of sampling. Diversity at both sites rose from May (496 & 565 BINs) to July (1,236 & 1,522 BINs) before decreasing in September (572 & 504 BINs). Unambiguous species names were assigned to 35% of the BINs (1,868) which represented 12,640 specimens. Another 7% of the BINs (386) with 1,988 specimens were assigned to genus, while 26% (1,390) with 12,092 specimens were only placed to a family. These results illustrate how a comprehensive DNA barcode reference library can identify unknown specimens, but also reveal how this potential is constrained by gaps in the quantity and quality of records in BOLD, especially for Hymenoptera and Diptera. As voucher specimens are available for morphological study, we invite taxonomic experts to assist in the identification of unnamed BINs.






