Abstracts GfBS-Tagung 2016
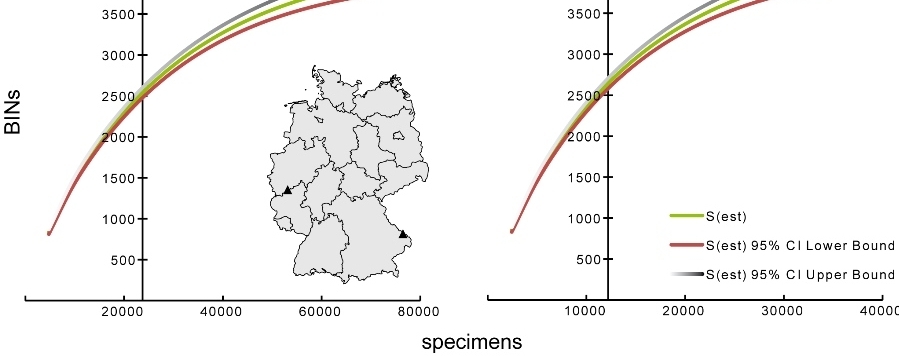
GBOL (www.bolgermany.de) is currently the largest barcoding campaign in Europe, financed by the German Federal Ministry for Education and Research. The initiative is flanked by complementing projects (Barcoding Fauna Bavarica (BFB), FREDIE for European freshwater fishes and invertebrate taxa, barcoding of the North Sea Fauna) and supported by more than 200 external experts. The latter play a crucial role, because the taxonomic workforce and expertise in natural history museums and universities is insufficient. Here, we present statistics and first results for the work achieved within the first three years (2011–2015) and give an outlook for the recently approved second funding period (2016–2018) introducing new partners and organism groups. Addressing the motto of this conference (Taxa in Time and Space) we also present first results from a cooperation with the Canadian Global Malaise Trap Program (GMP), where two collection localities in Germany were chosen for an annual sampling, in order to assess the diversity of flying arthropods. Applying the DNA barcode reference library we were able to assign species names to 35 % of the detected molecular units (5368 BINs) via reverse taxonomy. Furthermore, 7 and 26 % were assigned to genus and family level. These findings illustrate the potential of a comprehensive DNA barcode library to identify unknown samples, and reveal the gaps that need to be filled in the coming years. As the corresponding voucher specimens are available for morphological determination, we invite all taxonomic experts to explore the unnamed species and enhance the reference library by giving names to them.





