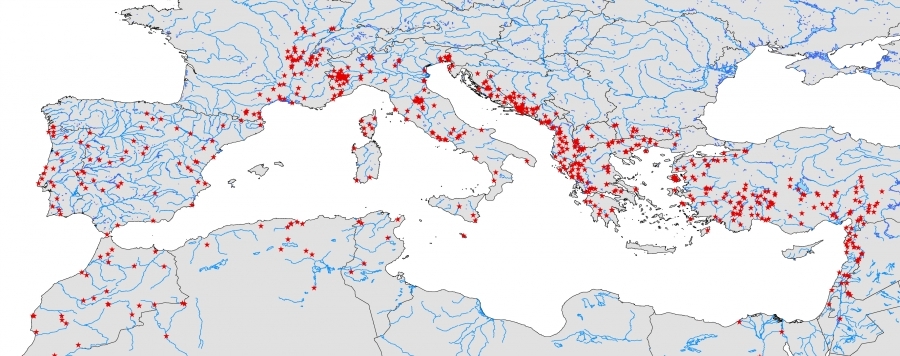
Fishes in the Mediterranean Biodiversity Hotspot
Incomplete knowledge of biodiversity remains a stumbling block for conservation planning and even occurs within globally important Biodiversity Hotspots (BH). Although technical advances have boosted the power of molecular biodiversity assessments, the link between DNA sequences and species and the analytics to discriminate entities remain crucial. Here, we present an analysis of the first DNA barcode library for the freshwater fish fauna of the Mediterranean BH (526 spp.), with virtually complete species coverage (498 spp., 98% extant species). In order to build an identification
system supporting conservation, we compared species determination by taxonomists to multiple clustering analyses of DNA barcodes for 3165 specimens. The congruence of barcode clusters with morphological determination was strongly dependent on the method of cluster delineation, but was highest with the general mixed Yule-coal scent (GMYC) model-based approach (83% of all species recovered as GMYC entity). Overall, genetic morphological discontinuities suggest the existence of up to 64 previously unrecognized candidate species. We found reduced identification accuracy
when using the entire DNA-barcode database, compared with analyses on databases for individual river catchments. This scale effect has important implications for barcoding assessments and suggests that fairly simple identification pipelines provide sufficient resolution in local applications. We calculated Evolutionarily Distinct and Globally Endangered scores in order to identify candidate species for conservation priority and argue that the evolutionary content of barcode data can be used to detect priority species for future IUCN assessments. We show that large-scale barcoding
inventories of complex biotas are feasible and contribute directly to the evaluation of conservation priorities.






